Molecular Assembly Modeling
Selected publications / opensource software
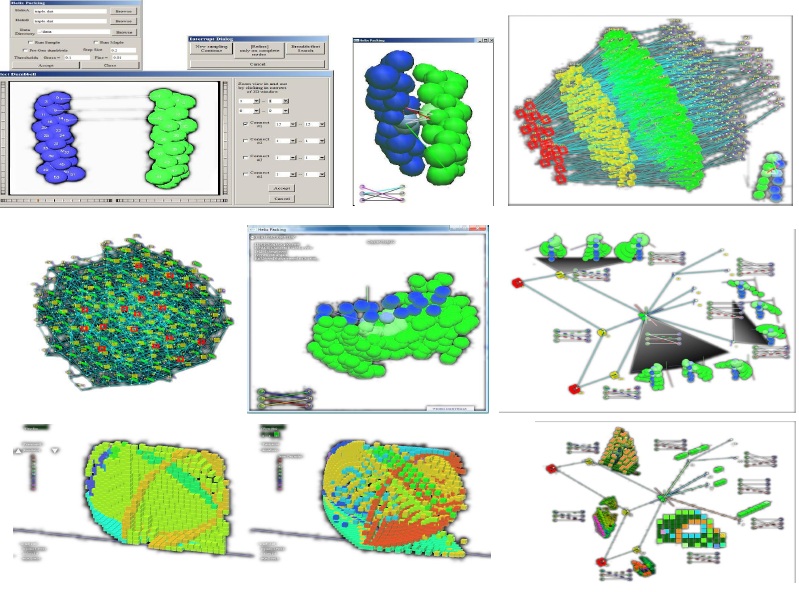
Supramolecular or macromolecular assembly is a remarkable phenomenon that occurs spontaneously and widely
in nature and underlies many processes, including
assembly of viral capsids and the action of drugs.
A molecular assembly configuration space refers to the set of configurations (positions and orientations)
that a collection of rigid molecules assume relative to each other,
in the presence of force fields, each of which is between a pair of atoms situated in different molecules.
The configuration space is typically high dimensional and geometrically intricate,
yet it determines configurational entropy, which is important in
determining the preferred assembly configurations of the molecular collection.
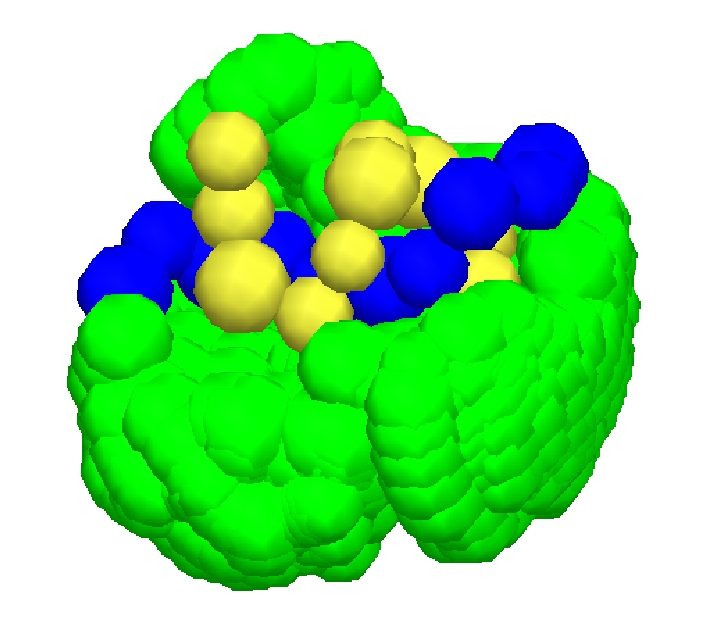 Predicting these preferred configurations or designing molecules that arrive at them
requires efficient and accurate free energy and configurational entropy computation,
which is a notoriously difficult problem that computational molecular scientists have studied for decades.
Established methods are generally based on full-blown molecular dynamics (MD) simulations or exhaustive Monte Carlo (MC) sampling of the configuration space.
Both methods suffer from the dual curses of dimensionality and geometric intricacy of the configuration space.
Predicting these preferred configurations or designing molecules that arrive at them
requires efficient and accurate free energy and configurational entropy computation,
which is a notoriously difficult problem that computational molecular scientists have studied for decades.
Established methods are generally based on full-blown molecular dynamics (MD) simulations or exhaustive Monte Carlo (MC) sampling of the configuration space.
Both methods suffer from the dual curses of dimensionality and geometric intricacy of the configuration space.
By modeling the pairwise intermolecular atomic force fields as a geometric constraint system, the Sitharam group has leveraged a theory of stratification and parametrization of assembly configuration spaces of geometric constraint systems developed by the group, and designed an efficient algorithm for obtaining an atlas of an assembly configuration space that captures the intricate geometry as interconnected regions of varying dimensions.
Aysegul Ozkan, a PhD student in the group developed
an opensource software EASAL
implementing these algorithms,
with help from James Pence, a previous undergraduate researcher in the group
and Ruijin Wu, a co-advised PhD student.
EASAL is currently being tested by a computational chemist (Maria Kurnikova of CMU) and physicist (Miranda Holmes-Cerfon, NYU) interested in using EASAL in conjunction with traditional methods for configurational entropy and free-energy computations.
EASAL's predictions of crucial intermolecular interactions that drive assembly of viral capsids
have been validated by a structural biology collaborator Mavis Agbandje-Mckenna at UF.
Another aspect of entropy that plays a crucial role in the case of symmetric macromolecular assemblies
such as the icosahedral viral capsid assembly is the notion of
combinatorial entropy that indicates the number of different pathways of subassembly formations that lead to successful assemblies.
In collaboration with Mathematicians Miklos Bona and Andy Vince at UF, the Sitharam group has given a method to explicitly count a simplified type of assembly pathways for symmetric macromolecular assemblies.